Feature-level Analysis
In feature-level data analysis for paired samples design, the major tasks include identifying (1) features that change over time, (2) features that are differenital between groups over time, and (3) features whose changes over time differ between groups. Features in (3) are a subset of features in (2). These tasks can be achieved by generate_taxa_test_single and generate_taxa_change_test_pair. The generate_taxa_test_pair function performs differential abundance analysis (DAA) based on compositional data using the mixed effects mode of LinDA, which is a DAA method based on log linear model with bias correction due to composiitonal effects. If the data are not compositional, standard linear mixed effects model (lme4) will be used.
Zhou H, He K, Chen J, Zhang X. LinDA: linear models for differential abundance analysis of microbiome compositional data. Genome Biol. 2022 Apr 14;23(1):95. doi: 10.1186/s13059-022-02655-5. PMID: 35421994; PMCID: PMC9012043.
Categorical vs Continuous Predictors
The generate_taxa_test_pair function automatically distinguishes between categorical and continuous predictor variables for the group.var parameter:
Categorical variables (factor/character): Creates pairwise comparisons vs. the reference level (e.g., "Treatment vs Placebo (Reference) [Main Effect]" and "[Interaction]" effects)
Continuous variables (numeric/integer): Tests linear association with the variable
This automatic detection allows the same function to handle both types of predictors appropriately.
Data Type Handling
In the generate_taxa_test_pair function, the feature.dat.type parameter specifies the data type in the feature matrix. Depending on the data type, different preprocessing steps and methods are used.
countandproportion: For count and proportion data, which are both compositional,lindawill be called. To address zeros, a pseudo-count of 0.5 is added to the count data, and zeros are substituted with half of the nonzero minimum for proportion data (feature-wise). Winorization at 97% quantile is performed to reduce the influence of outliers.other: Whenfeature.dat.typeis set to "other", the data are considered to be non-compositional and standard linear mixed effects model will be used. Users need to determine the appropriate normalization and transformation of the data before running the function. This option increases the applicability of MicrobiomeStat to other data types.
The prev.filter and abund.filter parameters filter taxa based on their prevalence and average relative abundance, so those rare and less abundant taxa will be excluded from testing. As the statistical power for these rare/less abundant taxa tends to be low, excluding them can reduce multiple testing burden.
The feature.level determines what aggregation level(s) the tests will be performed. If feature.level is set to "original", the original features in feature.tab will be tested. The feature.level can also be set to the aggregated levels specified in the feature.ann matrix.
The generate_taxa_test_pair outputs a table of LinDA association statistics for all tested taxa/features. The table contains the following components:
Variable: It identifies the taxon/feature being analyzed.Coefficient: Expressed as log2FoldChange, bias-corrected. It indicates the degree and direction of change in the abundance of a specific taxon/feature.SE: Standard errors of the coefficients. It measures the variability of the coefficient estimates.Mean Abundance: The average abundance of a particular taxon/feature across all samples.Prevalence: The proportion of samples where a specific taxon/feature is present.
The generate_taxa_change_test_pair function contains an additional parameter feature.change.func, which specifies the method or function used to compute the change between the two time points. The options include:
"absolute change"(default): Computes the absolute difference between the values at the two time points (value_time_2 - value_time_1)."log fold change": Computes the log2 fold change between the two time points."relative change": Computes the relative change as(value_time_2 - value_time_1) / (value_time_2 + value_time_1). If both time points have 0 values, the change is defined as 0.A custom function: If a user-defined function is provided to calculate the change, it should take two numeric vectors as input corresponding to the values at the two time points (
value_time_1andvalue_time_2) and return a numeric vector of the computed changes.
Results from both functions can be visualized using generate_taxa_volcano_single, will produce a volcano plot. It visualizes the relationship between the effect size (log foldchange) and its statistical significance. The function has the feature.sig.level and feature.mt.method parameters:
feature.sig.level: This parameter determines the significance level, influencing the position of the dashed lines in the volcano plot. It sets the threshold for distinguishing between significant and non-significant differences.feature.mt.method: Thi parameter determines whether the fdr-adjusted p-values or raw p-values will be plotted . There are two options available currently: "fdr" (false discovery rate) and "none" (raw p-value).
By understanding and appropriately setting these parameters, users can ensure a more accurate and contextually relevant interpretation of the plotted results.


For specific comparisons, labels like $Genus$Placebo vs LGG (Reference) [Main Effect] and $Genus$Placebo vs LGG (Reference) [Interaction] may appear.
The
[Main Effect]label indicates the primary difference in taxa abundance between the compared groups at the baseline.The
[Interaction]label reveals the interaction between group differences and time variable, i.e., whether the group difference changes with the time.
If the aim is to specifically investigate the shifts in taxa abundance between the two time points, the generate_taxa_change_test_pair() function can be used. This function employs a linear model (lm) to evaluate the changes in taxa abundance in relation to a grouping variable.

The results enable researchers to recognize taxa that exhibit notable changes in abundance across time points, and to determine whether these changes differ by group. In the following sections, we will demonstrate how to visualize taxa of interest such as those from the differential abundance analysis using various visualization tools.
First, let us look at generate_taxa_indiv_boxplot_long():

This function creates a series of boxplots, one for each taxon, and outputs them into a multi-page PDF. Several parameters in this function and other related functions are noteworthy.
feature.dat.type: One of "count", "proportion" or "other". For "count", the data will be converted to proportion data before the visualization.t0.level: the name of the baseline level ("t1") or the first time point.ts.levels: a character vector of the names of other levels to be visualized. In the paired samples setting,ts.levelscan be the name of the second time point ("t2")transform: This parameter indicates the transformation to apply to the abundance data when plotting. Transformations are only applied when thefeature.dat.typeis set to either "count" or "proportion". Whenfeature.dat.typeis "other", no transformation will be performed. User should deterimne the appropriate transformation to better visualize the data. The available options fortransforminclude:"identity": No transformation (default)"sqrt": Square root transformation"log": Logarithmic transformation. Zeros are replaced with half of the non-zero minimum for each taxon before log transformation.
feature.level: Specifiy which level of the data to be plotted.features.plot: This parameter can be used in all feature-level visualization functions to specify which taxa or features should be visualized. This is particularly useful for focusing on the results of differential abundance analyses. When you provide a vector of taxa or feature names tofeatures.plot, this will directly select these features for visualization, overriding any settings inprev.filterandabund.filter. By using this parameter, you can directly highlight and examine the taxa or features that are significantly different in abundance across your comparisons.top.k.plotandtop.k.func: In many scenarios, especially when navigating through expansive datasets, you may want to focus on a select subset of taxa that stand out either due to their high abundance or large variability.top.k.plotlets you visualize the top k taxa/features based on the criterion you selected intop.k.func, which have four choices:"mean": Highlights taxa with the highest average abundances across samples."sd": Selects taxa with the greatest variability (standard deviation) across samples, useful for identifying taxa with notable differences under varying conditions."prevalence": Chooses taxa with the highest occurrence across samples, targeting those most consistently present.top.k.funccan also accept a user-defined function so that the users can rank taxa based on their own criterion. The user-defined function should take the abundance matrix (those infeature.tabandfeature.agg.list) as the input and returns a numeric vector of the feature importance values. This allows for criteria beyond "mean", "sd", or "prevalence".
The generate_taxa_boxplot_long() places all taxa onto a single page, providing an overview of all your taxa at once.

Next, we present two key functions, generate_taxa_indiv_change_boxplot_pair() and generate_taxa_change_boxplot_pair(), for visualizing the changes from "t1" to "t2".


The generate_taxa_barplot_pair function will generate starked bar plots of taxa. In each sample pair, the same taxa are connected by lines ito track their abundance changes. The function outputs both individual and averaged bar plots. The function has an important parameter feature.number.
feature.number: This parameter determines the maximum number of taxa/features that will be visualized directly in the barplot. For datasets with numerous features, it's practical to limit to the most abundant or significant taxa, ensuring that the visualization remains informative and isn't cluttered. When the number of taxa surpasses the value defined infeature.number, the function aggregates low-abundance taxa into a collective category labeled "other". This means, for instance, if there are over 20 features in the dataset butfeature.numberis set to 20, the least abundant features that exceed this count will be collectively presented as "other" in the visualization. This approach ensures that the chart remains legible, highlighting the most dominant features, while still accounting for the contributions of less abundant taxa.


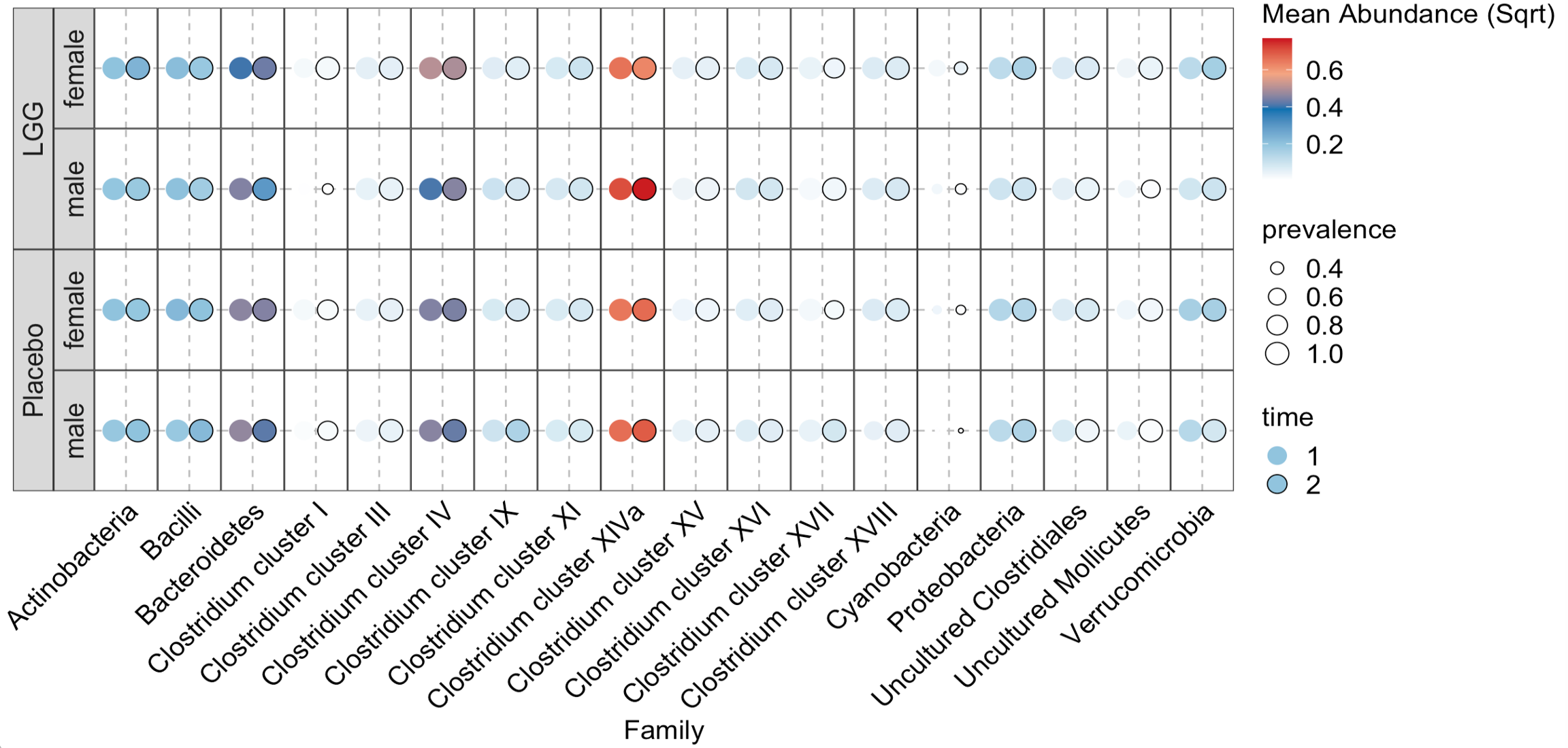
The function generate_taxa_dotplot_pair() depicts the mean abundance and prevalence across times and groups. The two time points are visualized together for easy comparison.
The generate_taxa_change_dotplot_pair() function visualizes the mean abundance change and prevalence change across groups:

The generate_taxa_heatmap_pair() function creates a heatmap of paired smaples. Here is an example:

Heatmaps are particularly insightful for discerning clusters of microbial families that exhibit similar abundance profiles across different samples. Heatmaps are particularly insightful for discerning clusters of taxa/features that exhibit similar abundance profiles across amples. By default, the function will only cluster rows (taxa/features) based on their abundance patterns. The columns are ordered by group.var and strata.var. If users wish to see the taxa in their original order without clustering, they can achieve this by setting cluster.rows = FALSE. The function also allows for column clustering by setting cluster.cols = TRUE, which can be instrumental in revealing groups of samples with similar abundance profiles. Note that when feature.dat.type is "count” or "proportion", the data will be visualized using proportions. For "other" type, the original scale will be used.
The generate_taxa_change_heatmap_pair() function helps assist in analyzing patterns of changes:

Scatterplot Visualizations
The generate_taxa_change_scatterplot_pair() function creates scatterplots showing the change in taxa abundance between two time points. This visualization is particularly useful for identifying taxa with large changes and understanding the direction of change across groups.
The feature.change.func parameter supports several methods for computing change:
"absolute change": Simple difference between time points"log fold change": Log2 fold change (with automatic zero imputation)"relative change": Relative change as (ts - t0) / (ts + t0)
For individual-level visualization, the generate_taxa_indiv_change_scatterplot_pair() function shows taxa changes for each subject:
These scatterplot functions complement the boxplot and heatmap visualizations by providing a different perspective on taxa abundance changes in paired samples.
Last updated